The art and science of ADC bioanalysis: a storyboard approach to assay design

In her latest column, Cathy comments on the ‘ever-raising bar’ posed by critical reagents and combines scientific practices with artistic visuals to make the most of ADC assay design.

Catherine Vrentas
Bioanalytical SME
Cathy Vrentas was most recently a Life Sciences Lead Associate at Booz Allen Hamilton (VA, USA), where she supported a large portfolio of federally-funded, preclinical through clinical programs in the CGT space and specializes in clinical trials and bioanalysis. Prior to this role, she was a Principal Scientist and managed a team of ~20 scientists at Thermo Fisher Scientific (VA, USA). In this role, she led the development and validation of 100+ assays to assess samples for preclinical programs and clinical trials for pharma and biotech, including work on multiple first-in-human trials for rare diseases and gene therapies. Cathy has experience in regulated immunoassays and cell-based assays for PK, ADA, NAb and biomarker applications, as well as enzymatic assays, oligonucleotide assessments, COVID-19 assays and tissue-based assessments.
Cathy received her BSc in Biochemistry and Molecular Biology from Penn State (PA, USA), a PhD in Cellular and Molecular Biology from the University of Wisconsin-Madison (WA, USA), an MBA from Longwood University (VA, USA), and an MPH in public health practice from Des Moines University (IA, USA). She has mentored over 60 scientists, students and summer interns in laboratory methods over her career and has volunteered for diverse nonprofits including the foster care system, dementia education, prison education, youth science outreach, public health, oyster restoration and health advocacy.
Every ligand-binding assay (LBA) developer in our midst likely has a story (or two!) involving a problematic critical reagent. Perhaps the culprit was a capture-detection reagent pair that just wasn’t selective enough to perform well with patient matrix samples or a new lot of an antibody that was not comparable to the previous lot. I’ve also personally been victim to labeled reagents that aggregated or degraded more rapidly than expected, necessitating screens of buffer compositions and storage conditions.
Excellent assays start with consistent reference standards and appropriate, specific, selective and stable key materials. The impact of critical reagent selection on bioanalytical project management is amplified by the time it takes to produce new batches of materials, especially new antibodies; a developer who must go back to the drawing board could lose months as new reagent options are generated and screened.
The reagent bar is raised even further for the assay developer working on a therapeutic in the ADC (antibody-drug conjugate) class. ADCs combine the promise of the target specificity of an immunotherapy with the cancer cell-killing potential of a potent toxin. For those who are newer to working with ADCs, this Bioanalysis Zone Spotlight on ADC bioanalytics explores the relevant fundamentals, such as techniques for separately quantitating levels of multiple components of the therapeutic in patient biofluid samples, including the fully conjugated therapeutic, free toxin/payload that has been liberated from the antibody, and unconjugated antibody.
With the need for multiple assay types typically comes the need for a whole suite of critical reagents. Program teams may choose to use bioanalytical assays to measure, for example, not only the three therapeutic forms and components listed above, but also total antibody and the levels of metabolites of the free or conjugated drug payload, in order to characterize the exposure of the body to the drug. Custom reagents must then be carefully screened to ensure that they have the appropriate specificity, whether they are binding an idiotype on the antibody or the intact or metabolized toxin. Indeed, bioanalytical approaches for ADC quantitation typically use a mixture of LBA and mass spectrometry approaches to provide the flexibility required to assess the range of component sizes and modifications, and some molecular forms are challenging to detect with available LBA critical reagents. Ligand binding assay specialists are likely to be involved in generation of critical reagents for ADA and NAb assays for ADCs as well.
Due to the complexity of the suite of bioanalytical assays involved, ADCs are a perfect opportunity to adopt a modular, graphical approach to visualize how different critical reagents can be arranged to create an appropriate and optimized assay design. Multiple formats are possible:
- Develop a PowerPoint slide with a standardized set of images to represent molecules involved in the assays for the program, including different forms of ADCs, target and antibody reagents. Pre-made graphics for public use (with appropriate citations) can be obtained through sites like the NIH BioArt webpage, which provides free antibody images that can be downloaded in a spectrum of colors: Bioart
- Go a step beyond a hand-sketched diagram and create a custom set of magnets representing the different critical reagents. These can be easily and cheaply made with magnetic sheets (some of which are inkjet printable) for training purposes. Your team can use these tools to rapidly rearrange assay design combinations and predict the impact of interfering molecules.
Assay design toolkit

Figure 1: Colorful components of your assay design kit, including molecules representing antibodies and protein targets, can be rearranged and reused across designs and serve as excellent and training tools.
Of course, these assay “storyboards” do not have to be limited to use for ADC applications; they provide an opportunity for active learning for new (as well as more seasoned) method developers, validators and users across a range of LBA types and modalities. Graphical depictions can incorporate visualizations of target binding, dimerization of antibodies or targets and/or interactions with other interfering matrix proteins to inform design.
Bioanalytical scientists should also plan to map out the biochemistry of the ADC at a finer level to understand the expected sites of payload conjugation, the chemistry of conjugation and the expected heterogeneity of the drug-antibody conjugation ratio (DAR) of the product. For example, is the payload conjugated to specific, engineered sites in the antibody, or conjugated more stochastically to available -NH2 groups on lysine amino acids? Especially if high DARs are expected, the conjugated payloads could cause steric hindrance and impact critical reagent binding. Bioanalytical scientists with experience in biotin conjugation or ruthenylation of reagents for other assay formats will be familiar with many related concepts and can apply this knowledge to their interactions with CMC (chemistry, manufacturing and control) teams to learn more about the biochemistry of the ADC molecule.
Unlike a standard monoclonal antibody drug reference material, an ADC may be heterogeneous even before it enters the body, with molecules representing a range of DARs. Therefore, the bioanalytical scientist must:
(a) determine what forms of the ADC are needed as reference material
(b) make plans to test the impact of different DARs and/or metabolized products on the performance of each assay as needed for the state of development of the product
(c) ensure that the assays are designed to measure the intended population of molecules.
Storyboards can be used to visualize the predicted impact of a particular assay format or critical reagent selection on analyte measurement. For example, the use of an anti-payload antibody for detection in an ELISA can create variability in signal across ADCs of different DARs, as more detection antibodies would be predicted to bind per high DAR ADC than to a low DAR ADC. In contrast, capture with the same anti-payload antibody would be expected to improve consistency of binding across ADCs of different DARs [1,2]. Therefore, the design of the assay must be matched to the ultimate bioanalytical goals.
Using your toolkit for ADC assay design visualization
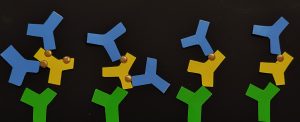
Figure 2: Depicted here is the effect of capture and detection antibody selection on the DAR dependence of your assay, as discussed and graphically depicted in Kumar et al. (2015) [1]. In green, capture antibodies bind to the protein backbone of the ADC candidate (depicted in yellow). Detection antibodies (blue) bind to the payload of the ADCs (gold); when the ADCs have higher DAR, there may be a difference in signal generated from the ADC at a higher DAR (left side of image) vs a lower DAR (right side of image) due to the increase in payload sites available for binding per molecule. Here, the payload was depicted using gold stickers that were added to each ADC component.
Bioanalytical assay designs will also need to continue to adapt to the advances in the ADC field, which fuse overall trends in large-molecule drug design with the ADC platform. For example, new conjugation strategies allow for antibodies to be linked to multiple toxin payloads, and payload types and activities continue to diversify. Targeting to solid tumors (via binding to proteins specific to the tumor microenvironment) remains of high interest, as does utilization of ADC therapies for non-oncologic applications like immunomodulation. By starting with the fundamentals of critical reagent planning and evaluation and combining that with analysis of assay design from the early stages of a project, LBA-focused bioanalytical scientists will be well-equipped to tackle the ever-increasing assay complexity.
Disclaimer: the opinions expressed are solely those of the author and do not express the views or opinions of Bioanalysis Zone or Taylor & Francis Group.