More bang for your buck: newly developed RNA-based sensor detects protein biomarkers for USD $1
Protein biomarkers are useful indicators of chronic or acute conditions and can be used to detect arthritis, cancer, Alzheimer’s disease, PTSD and a whole host of other conditions. Conventional testing of these biomarkers can cost USD $100-1000 or, in some cases, even more. To combat this issue, Penn State University (PA, USA) researchers have developed a low-cost, RNA-based technology to detect and measure protein biomarkers. The results of the study were published in Nature Communications.
The team at Penn State comprised of Howard Salis, Associate Professor of Biological, Chemical and Biomedical Engineering, Grace Vezeau, who earned her doctorate in Biological Engineering from Penn State in 2021 and Lipika Gadila, who earned her BSc in Chemical Engineering from Penn State in 2018.
The team demonstrated that RNA-based sensors can be engineered to detect protein biomarkers. These included monomeric C-reactive protein, which is used to monitor chronic inflammatory conditions such as heart disease and arthritis, as well as interleukin-32 gamma, a signaling protein associated with acute viral or bacterial infections. Salis explained how these RNA-based sensors could be used to develop devices for diagnostic testing:
“These tests can help a clinician diagnose a patient, but it’s more informative to carry out multiple biomarker measurements periodically over the span of several weeks. Right now, one test can be expensive, and they add up. With our new RNA-based technology, it’s now possible to carry out the same measurements for much less.”
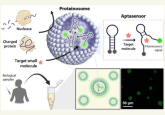
The sensor is comprised of two elements. The first is a cell-free expression system, which contains cellular machinery to read DNA and produce proteins without the restriction of cell membranes, allowing ‘bulky’ proteins to enter freely. The second element consists of RNA-based sensors called riboswitches, which are engineered to bind to a biomarker protein and regulate activation or suppression of an observable signal. The riboswitch is formed inside the cell-free expression system using instructions from DNA. Both elements together cost approximately USD $1 per reaction.
This is the first study whereby researchers have engineered a riboswitch sensor to detect biomarker proteins. However, the challenge lies in deciphering the best DNA instructions to produce protein sensors with maximum sensitivity:
“Past efforts to engineer such riboswitch sensors have largely relied on trial-and-error experimentation, for example, constructing and characterizing large random libraries to identify riboswitch variants — the genetic blueprints and aptamers — that work best” continued Salis. Using a combination of thermodynamic modeling and computational optimization, we rationally designed new riboswitches that are predicted to be excellent protein sensors and then we tested them. Our design algorithm is called the Riboswitch Calculator.”
Salis and the research team tested their new technology by detecting three proteins: MS2 as a proof-of-principle test, as well as human monomeric C-reactive protein and human interleukin-32 gamma, two medically relevant biomarkers. The researchers engineered 32 riboswitches, the majority of which successfully sensed their target proteins.
“Current assays require expensive detection reagents, expensive and bulky instruments, sample cold chain storage and distribution, and trained personnel” Salis stated. “By applying modeling and computational design, we engineered low-cost protein sensors that can be freeze-dried and rehydrated. The next step is to develop an easy-to-use device that allows researchers and clinicians to use this new technology.”
Reference: Vezeau GE, Gadila LR & Salis HM. Automated design of protein-binding riboswitches for sensing human biomarkers in a cell-free expression system. Nat Commun. 14(1):2416. doi: 10.1038/s41467-023-38098-0 (2023)