End of the road? The future of chromatography

Isabelle Kohler studied Pharmacy at the University of Geneva, Switzerland. She carried out her PhD at the School of Pharmaceutical sciences, University of Geneva, and obtained her PhD in Pharmaceutical Sciences in 2013, focusing on the use of capillary electrophoresis hyphenated to mass spectrometry in clinical and forensic toxicology. She moved to The Netherlands for a postdoctoral fellowship carried out at the Leiden University Medical Center in the Center for Proteomics and Metabolomics, where she investigated the biomolecular mechanisms of familial hemiplegic migraine in a transgenic mouse model using untargeted and targeted metabolomics approaches. From July 2016 to May 2020, she worked as Assistant Professor in the group of Analytical Biosciences and Metabolomics at the Leiden Academic Center for Drug Research, Leiden University. Her research interests include clinical metabolomics, brain metabolism, neurodegenerative disorders and neurological diseases, method development and validation, as well as data analysis and data integration.
Correspondence: Dr Isabelle Kohler, Bioanalytical Chemistry, Amsterdam Institute for Molecular and Life Sciences, Vrije Univerisiteit Amsterdam, The Netherlands. Email: [email protected]
Mass spectrometry in bioanalysis
Mass spectrometry (MS) has become the gold standard analytical technique in a large diversity of bioanalytical applications. Over the last decades, many technological developments have led to the commercialization of ‘horse race’ instruments capable of outstanding performance. One of the most relevant breakthroughs was certainly the advent of high-resolution mass spectrometry (HRMS), pushing the boundaries of resolving power and mass accuracy, which are both essential in bioanalysis for the identification and characterization of unknown compounds. Mass analyzers that can perform HRMS include time-of-flight (TOF), Orbitrap and Fourier transform ion cyclotron resonance (FT-ICR), which typically offer mass accuracies lower than 5 ppm and can reach mass resolutions up to 1,000,000 for FT-ICR-MS instruments at m/z 400. Table 1 highlights the differences between the three instruments in terms of mass accuracy, resolving power, sensitivity, speed of analysis and costs.
Table 1. Comparison between TOF, Orbitrap and FT-ICR instruments [1].
|
Mass analyzer |
Mass accuracy | Mass resolution | Sensitivity | Speed | Costs |
| TOF | ++ (5 ppm) | + (50,000) | +++ | +++ | +++ |
| Orbitrap | +++ (< 5 ppm) | ++ (500,000) | ++ | ++ | ++ |
| FT-ICR | ++++ (< 1–2 ppm) | +++ (1,000,000) | ++ | + | + |
Those instruments also enable tandem mass spectrometry (MS/MS) experiments by adding a fragmentation step based on collision-induced dissociation, electron-capture dissociation or infrared multiphoton dissociation, which offer additional confidence in identification and in-depth characterization of unknown compounds [2]. One of the latest developments in this field is SWATH technology, which stands for Sequential Window Acquisition on all THeoretical fragment ion spectra and is implemented on some commercially available quadrupole-time-of-flight (qTOF) and q-Orbitrap instruments. SWATH-based approaches rely on a data-independent acquisition using narrow isolation windows, where all detected precursor ions are sequentially fragmented in a series of quadrupole isolation windows, allowing for both qualitative and quantitative analysis of a large number of analytes in biosamples [3].
All these technological developments have fostered the use of direct infusion mass spectrometry (DIMS) and flow injection analysis-mass spectrometry (FIA-MS) approaches in bioanalysis, where the sample is directly infused or injected into the mass spectrometer after adequate sample preparation and without any additional chromatographic or electrophoretic separation. The major advantage of DIMS and FIA-MS approaches is the possibility for high-throughput analysis, reaching analysis times as short as 1–2 min. As an example, DIMS is very popular in lipidomics, in an approach known as shotgun lipidomics. In shotgun lipidomics, lipids are first extracted using selective sample preparation techniques prior to selective ionization, i.e. the intra-source separation of the different lipid classes based on their distinctive structures. This strategy, combined with the continuous infusion of the prepared sample into the mass spectrometer and multiple acquisitions in different scan modes, enables the qualitative and quantitative analysis of a large diversity of lipids without the need for chromatographic or electrophoretic separation [1].
The excellent qualitative and quantitative performance obtained with state-of-the-art HRMS instruments might therefore make the bioanalytical scientist doubt the usefulness of a separation step prior to the MS detection. Indeed, why would one still need to use chromatography (or electrophoresis) in bioanalysis, considering the substantial added time for both the method development and the analysis itself – not to mention the added overall costs?
The complexity of the human metabolome
The fourth version of the Human Metabolome Database (HMDB), introduced in 2017, reported more than 110,000 fully annotated metabolites – a remarkable number, considering that it only takes into account compounds with a mass lower than 1,000 Da. The human metabolome encompasses a large diversity of metabolites including amino acids, lipids, organic acids, carbohydrates, small peptides, hormones, vitamins and drug metabolites, which makes its comprehensive analysis very challenging – if not currently impossible. Moreover, many of those metabolites present very similar physico-chemical properties, requiring analytical approaches to enable their separation. Examples of such compounds include isomers, typically lipid isomers or optical isomers such as enantiomers, which usually have very different physiological activities [4].
An excellent example to illustrate this is 2-hydroxyglutarate (2-HG), which is the first oncometabolite ever reported. 2-HG is a metabolite produced upon gain-of-function mutations of the enzymes isocitrate dehydrogenase 1 and 2 (IDH1/2), competing with α-ketoglutarate in multiple cellular processes including DNA methylation, histone demethylation and epigenetic modeling, with the overall effect of priming cells for malignancy [5]. The 2-HG molecule carries an asymmetric carbon atom in its carbon backbone and therefore exists in two distinct enantiomers, L-2-hydroxyglutarate (L-2-HG) and D-2-hydroxyglutarate (D-2-HG). Both L-2-HG and D-2-HG are normal endogenous metabolites present in all human body fluids, but D-2-HG is also produced as a result of mutations of IDH1/2 enzymes. L-2-HG is not affected by these mutations. In healthy individuals, endogenous serum levels of L-2-HG are equal to or exceed levels of D-2-HG. Overall, this shows the need for analytical methods that enable the discrimination between both enantiomers to avoid false-positive results [6].
Another example can be retrieved from lipidomics, when targeting bioactive lipids. Conventional MS and MS/MS approaches are not capable of separating lipid isomers that differ only in the position of the acyl chain or the double bond, or lipid isomers with a different geometry. 5S,12S-dihydroxy-6E,8Z,10E,14Z-eicosatetraenoic acid (5S,12S-DiHETE) and leukotriene B4 (LTB4, systematic name 5S,12R-dihydroxy-6Z,8E,10E,14Z-eicosatetraenoic acid [5S,12R-DiHETE]) are two diastereoisomers and geometrical isomers, as illustrated in Figure 1.
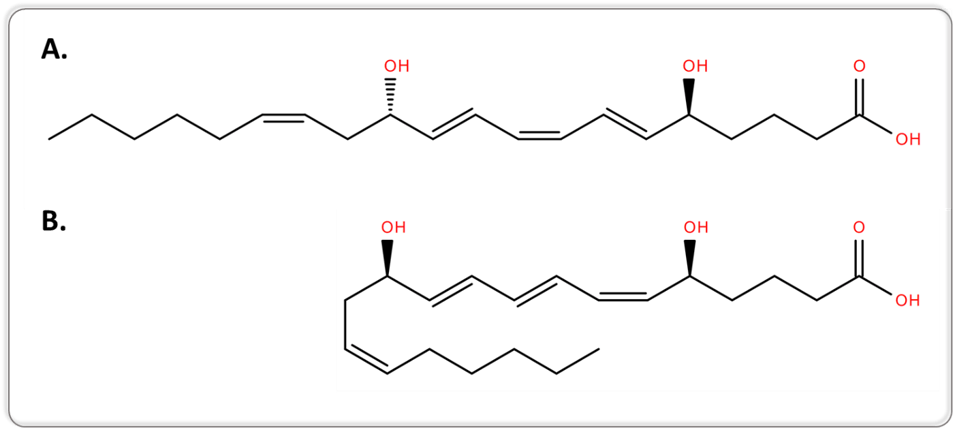
Figure 1.Two-dimensional structures of 5S,12S-DiHETE (A.) and LTB4 (B.).
5S,12S-DiHETE and LTB4 arise from different biochemical pathways and have different biological activities. LTB4 is an enzymatically-generated chemotactic lipid mediator, while 5S,12S-DiHETE is produced upon platelet–neutrophil interaction [7]. The separation of 5S,12S-DiHETE and LTB4 is therefore required when investigating the biological role of bioactive lipids to avoid misinterpretation of the results.
Do we need chromatography?
Even with the highest resolving power and mass accuracy, optical isomers cannot be separated using conventional MS and MS/MS approaches, as their exact mass and fragmentation pattern are both identical. Separation between such isomers, as well as between closely-related compounds, therefore relies on the addition of a separation step prior to the MS detection. This step does not only help in improving the discrimination between the target analytes, but also in separating the compounds of interest from the matrix interferents. Indeed, DIMS and FIA-MS approaches typically suffer from matrix effects, for example, ion suppression or enhancement, which can significantly affect the accuracy of the method. Matrix effects can be particularly relevant with complex matrices such as blood (plasma/serum), urine or tissue samples where lipids and salts can strongly impact the ionization efficiency.
In many cases, the addition of a separation step prior to the MS detection is thus beneficial, if not essential. Liquid chromatography (LC), especially reversed-phase liquid chromatography, is now considered the chromatographic technique of choice in bioanalysis, offering high selectivity, sensitivity and throughput when combined with MS. The current superiority of LC–MS over other separation techniques such as GC and CE is likely explained by the advent of columns equipped with sub-2-µm particles (ultra-high pressure liquid chromatography), which allows for dramatic improvements in resolution and throughput compared to the conventional HPLC format.
The last 2 decades have also seen tremendous technological innovations in the field of liquid-based separation techniques, such as hydrophilic interaction chromatography (HILIC), supercritical fluid chromatography (SFC), ultra-high performance supercritical fluid chromatography (UHPSFC), multidimensional LC, as well as pillar array columns to replace columns packed with spherical particles. This large and diverse portfolio offers a virtually infinite choice of chromatographic techniques enabling the discrimination of analytes, which would otherwise not be separated using DIMS or FIA-MS approaches. Diastereomers and geometrical isomers are then relatively easily separated using chromatography, while enantiomers can be analyzed using a chiral stationary phase in LC and SFC, or a chiral selector in CE. One of the latest developments in the field of liquid-based separation techniques, the use of micro-pillar array columns, has also shown the ability to enable the baseline separation of positional lipid isomers [8].
The special case of ion mobility – mass spectrometry
Ion mobility–mass spectrometry (IM-MS) is a recent technique, which appears to be very promising in a large diversity of bioanalytical applications. IM-MS adds an orthogonal separation dimension between the chromatographic separation step and the MS detection step without impacting the analysis time, as this extra separation step occurs in a timescale of milliseconds. In IM-MS, ions are separated according to their ion mobility (also called drift time) in a cell under an electric field in the presence of an inert buffer gas. With this drift time being intrinsically linked to the size, shape and charge of an ion, closely related compounds can be putatively separated. The two geometrical lipid isomers 5S,12S-DiHETE and LTB4 have been shown to be baseline separated using differential-mobility spectrometry, one of the commercially available IM-MS technologies, with two different compensation voltages [7]. IM-MS has also enabled the separation of two other biologically relevant lipid isomeric substances, the two protectins PD1 and PDX [7].
IM-MS also offers the advantage of being able to be directly combined with chromatographic and electrophoretic techniques, bringing an additional orthogonal dimension for discriminating between closely related compounds. Despite its relative novelty, IM-MS has been already used in many bioanalytical applications and will likely see an exponential growth in the coming years.
Conclusions and perspectives
Despite the tremendous technological developments carried out in the field of MS over the last years, a separation step prior to the MS detection remains beneficial, if not essential in some applications. This is particularly relevant for the analysis of isomers (geometrical isomers or enantiomers) which cannot be separated using direct infusion MS or MS/MS approaches. Moreover, the complexity of biological matrices also often requires the addition of a separation step to lower the risks of matrix effects and inaccurate results, even with adequate sample preparation.
For all the reasons discussed here, chromatography and electrophoresis still have a bright future. Liquid-phase chromatography has also seen multiple technological developments over the last 2 decades including UHPLC, SFC, multidimensional LC and pillar array columns which will certainly help LC hold its current position in the field of bioanalysis.
Finally, due to the advantages offered by adding an orthogonal separation step between chromatography and MS detection, IM-MS will probably become a key-player in bioanalysis in a very near future.
[1] Wang J, Wang C, Han X. Tutorial on lipidomics. Chim. Acta. 1061, 28–41 (2019).
[2] Ghaste M, Mistrik R, Shulaev V. Applications of fourier transform ion cyclotron resonance (FT-ICR) and orbitrap based high resolution mass spectrometry in metabolomics and lipidomics. J. Mol. Sci. 17(6), 816–838 (2016).
[3] Bonner R, Hopfgartner G. SWATH acquisition mode for drug metabolism and metabolomics investigations. Bioanalysis 8(16), 1735–1750 (2016).
[4] Haselberg R, Pirok BWJ, Gargano AFG, Kohler I. Clinical metabolomics: expanding the metabolome coverage using advanced analytical techniques. LC GC 32(9), 465–480 (2019).
[5] Yang M, Soga T, Pollard PJ, Adam J. The emerging role of fumarate as an oncometabolite. Front Oncol. 2(85), (2012).
[6] Struys EA. 2-Hydroxyglutarate is not a metabolite; D-2-hydroxyglutarate and L-2-hydroxyglutarate are! Natl. Acad. Sci. 110(51), E4939 (2013).
[7] Jonasdottir HS, Papan C, Fabritz S et al. Differential mobility separation of leukotrienes and protectins. Chem. 87(10), 5036–5040 (2015).
[8] Sandra K, Vandenbussche J, t’Kindt R et al. Evaluation of micro-pillar array columns combined with high resolution mass spectrometry for lipidomics. LC GC Eur. 30(6), 6–13 (2017).
Disclaimer
The opinions expressed in this interview are those of the author and do not necessarily reflect the views of Bioanalysis Zone or Future Science Group.
Our expert opinion collection provides you with in-depth articles written by authors from across the field of bioanalysis. Our expert opinions are perfect for those wanting a comprehensive, written review of a topic or looking for perspective pieces from our regular contributors.
See an article that catches your eye? Read any of our articles below for free.