Complexity of human diseases should promote the use of high-resolution mass spectrometry for routine untargeted analyses
Bertrand Rochat – University of Lausanne (Switzerland)
Bertrand Rochat obtained his PhD at the University of Lausanne in Switzerland. After various post-doc periods in France, the United States and Scotland, he worked 2 years as Laboratory Head at Novartis Pharma (Basel, Switzerland). From 2003 to 2017, he was in charge of the Quantitative Mass Spectrometry Facility at the University Hospital of Lausanne (Switzerland). In 2018, he moved to the University of Lausanne to be in charge of the peptidomics analysis.
Bertrand has published more than 50 original articles, editorials, book chapters and reviews in the following fields: routine and quantitative analysis using LC–HRMS or LC-QQQ-MS; implementation of various omics analysis in patient’s plasma; intratumoral drug metabolism and disposition; and determination of native peptides by LC–MS in biological matrices. He is also a consultant for Cormedica Corporation (France).
Key words: clinical, diagnostics, diagnosis, high-resolution, laboratory developed tests, liquid chromatography, mass spectrometry
Short overview of mass spectrometry for the last 2 decades
For the last 2 decades, the number of peer-reviewed articles found in the Pubmed database about mass spectrometry (MS) and high-resolution MS (HRMS) have been constantly increasing. Figure 1 shows the increase in the number of publications containing ‘mass spectrometry’ or ‘high-resolution mass spectrometry’ keywords. The last 20 years have seen 6- and 21-fold increases, respectively and 1.5- and 4-fold increases when only the last 10 years are considered. These trends are confirmed when other keywords such as ‘method validation’, ‘quantitative methods’ or ‘clinical’ are associated to MS and HRMS (Figure 1). The market of mass spectrometry (instruments sold) is still expected to increase over the next 2 decades [1].
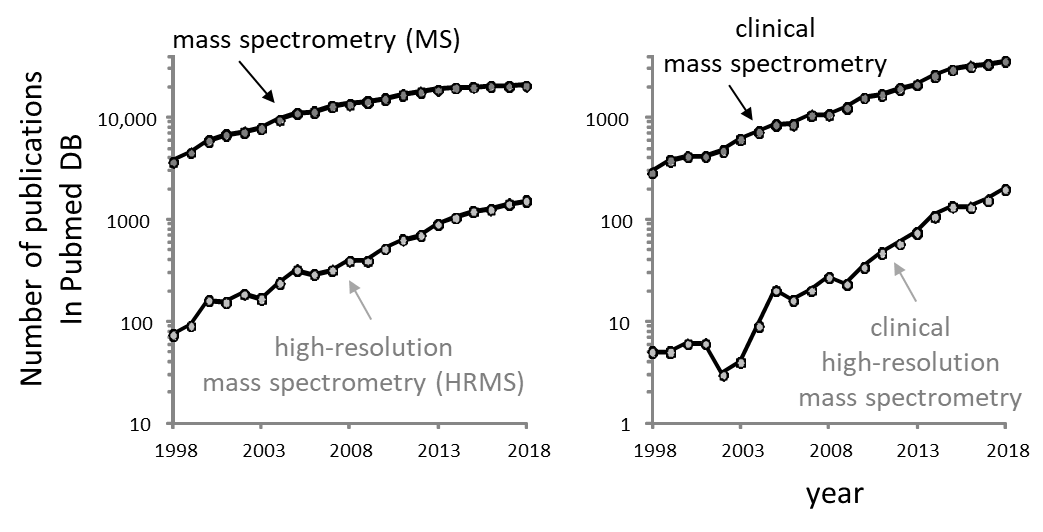
Interestingly, although HRMS publications are constantly increasing, they only represent a small fraction of the articles published about MS. Even if the keyword does not collect all articles using HRMS, this difference would be probably much bigger if the number of analyses performed in routine labs were considered. In fact, most routine analyses are not published and therefore, invisible to the PubMed database (the exceptions are clinical studies, case reports, etc.). From these observations, we could simply think that HRMS instruments are not made for routine GC–MS or LC–MS analyses – which are mostly performed by clinical, food safety, environmental and forensic labs – but rather that they are dedicated for research.
In clinical, environmental, forensic and food safety labs, bioanalysts have always compared the quantitative performance of various MS technologies. Two decades ago, HRMS was clearly inferior to many other MS instruments for quantitative and routine determinations. Therefore, at that time, HRMS was ‘reduced’ to accurate mass determination, structure elucidation, chemical formula identification and discovery. With the arrival of new HRMS technologies, mainly the orbitrap™ cell and the analog-to-digital converter for TOF-MS, a new start was launched for HRMS quantification. Therefore, it was suddenly very pertinent to carry out a side by side comparison of the quantitative performance of HRMS and its main MS competitor for routine analyses, the so-called, triple-quadrupole MS (QQQ-MS).
The inherited mindset and some facts strengthened the beliefs that HRMS instruments were: more complex, more expensive, less robust, less user-friendly, less selective in full scan, less quantitative and with a bigger footprint compared with QQQ-MS. Therefore, at the time there was little expectation to see HRMS becoming a major player in routine analysis.
However, most of these beliefs have been shown to be completely, essentially or at least partially wrong. For instance, HRMS is more selective, as user friendly and no more complex to use. In general, HRMS instruments are more expensive than QQQ-MS but the difference can be less than 25% when comparing similar MS instruments and not the cheapest QQQ-MS with, for instance, a tribrid™-orbitrap-MS (three mass analyzers in one instrument). MS manufacturers have commercialized cheaper HRMS instruments in addition to expensive ones that are dedicated to research investigations performed in the best academic laboratories. It is also true that some QTOF-MS instruments have a rather big footprint, preventing the ability to have many of these instruments in a small room. But this is not the case for many HRMS, e.g. Q-Exactive™ series from Thermo Fisher Scientific (MA, USA) and QTOF X500 series from SCIEX (MA, USA).
Some articles that have compared LC–MS analyses side by side with various instruments have described QQQ-MS as having better limits of detection. Although these comparisons always depend on the compounds and instruments used, generally the difference in sensitivity is small, in the region of two to five fold (for a comprehensive review see [2]). In general, the differences in sensitivity are inconsequential for most routine analysis because the compound analyzed is present in much higher levels than the limit of quantification of the MS equipment. Grund et al. [3] compiled eight peer-reviewed articles that compared HRMS and QQQ-MS quantitative performance. These eight articles show that the HRMS sensitivity was less than, equal to and greater than QQQ-MS for 19/108 (18%), 60/108 (60%) and 24/68 (22%) of compounds, respectively. Also, calibration linearity can show the same order of magnitude between both QQQ-MS and HRMS (e.g. 0.0125 to 5000 ng/mL in plasma extracts) [3]. These articles, and many others [4], show that HRMS and QQQ-MS are fully comparable for quantitative determinations, thus challenging the inherited mindset with regards to HRMS.
For almost a decade, my colleagues and I have compared the performance of HRMS and QQQ-MS in quantitative and routine analyses. In my in-depth studies, I have come to the conclusion that the HRMS instruments that we tested performed similarly to QQQ-MS for all aspects of routine, quantitative and targeted determinations [2,5]. The cherry on the cake was the additional capability of HRMS to perform all kinds of research and untargeted investigations. I tried to share these advantages to QQQ-MS users, but in many cases, I have found skepticism, poor interest and lack of motivation to change. I had to ask myself why there was such a gap between my understanding of HRMS capabilities and my vision for future biomedical analysis and the resistance of most bioanalysts working with QQQ-MS.
Although I could have been slightly too enthusiastic, my comparison between the two technologies was robust and took into account all aspects of clinical constraints. I was – and still am – sure that HRMS will bring a new paradigm to most routine labs and even clinical labs that are less inclined to perform screening analyses.
A few years ago, I knew that HRMS would bring the ability to do much more than just targeted and quantitative determinations since HRMS can perform various investigations on top of routine workflows. However, I believe that some bioanalysts do not fully appreciate the additional research capabilities brought by HMRS.
I was also not fully cognizant of the reality that HRMS will be needed to perform untargeted analysis within a routine frame. Even if this was rather obvious in ‘screening labs’ such as for the environment, food safety and forensic fields, this will also become true for diagnostics in clinical labs. The future need of untargeted routine analyses makes a big difference and promotes the use of HRMS due to its capabilities.
I can concede that there is possibly ‘some’ gap in favor of QQQ-MS for ‘simple’ quantitative determinations (e.g. ultrafast polarity switching and ultimate sensitivity with the most expensive QQQ-MS). But today, this gap is inconsequential for the great majority of analyses performed routinely. Furthermore, the cost per sample is almost identical for QQQ-MS and HRMS, estimated to be less than 5% difference.
But, one of the biggest gaps that prevents QQQ-MS users from buying HRMS for their routine analysis, is conservatism, poor interest and the lack of energy to activate the transition. Indeed, many lab heads working with routine and quantitative methods have been trained on QQQ-MS, which makes it ‘difficult’ to learn and adapt their knowledge to HRMS. Also, why should they rewrite their standard operating procedures for HRMS? Why should they change their way of working for a technology that does not surpass QQQ-MS for quantitative and routine analyses?
I believe HRMS should be adapted as soon medical doctors will be asking for untargeted analysis for more global diagnostic screenings. The biomedical need for untargeted screening should force bioanalysts to purchase adequate instruments, for example, HRMS that can allow fast scan capabilities and selective detection by accurate mass determinations. As a consequence, MS manufacturers will adapt to users’ choice and will develop cheaper HRMS instruments, highly adapted to routine quantitative analysis. This will be done in addition to the very expensive and sophisticated HRMS equipment, which is dedicated to research (Tribrid™-MS, ion-mobility QTOF-MS etc.).
MS manufacturers, to some extent, have already felt this shift. Indeed, this shift can be observed with the use of HRMS for performing routine analysis of biologics such as therapeutic antibodies. Although MS manufacturers still strongly promote QQQ-MS for quantitative analyses (possibly as the result of the current organization of production lines), they have already developed cheaper quantitative HRMS. This evolution should go further because of the need for more global untargeted routine analyses – we need to see a more complete picture.
What is untargeted routine analysis and why should we perform it?
We all know that living systems are complex, but we have probably underestimated their degree of complexity and diversity. For a few centuries, scientists have looked at biological systems and sciences in general with a reductionist outlook/paradigm. We have searched for and discovered biomarkers based on this segmented and simplified approach. This road has been successful and today, many biomarkers are determined in clinical labs. Despite this success, our ability to discover new biomarkers is now a real concern [6–9]. Indeed, even though discoveries of new potential biomarkers are published every month, the overwhelming majority of them do not enter the clinical laboratory. This ‘hangover’ in biomarker discovery is particularly bitter because, today more than ever, scientists have various ultra-performing technologies capable of collecting big amounts of data (e.g. LC–MS or high-throughput DNA sequencing analyses) at their disposal.
There are many explanations for the difficulty to transfer research findings to the clinic. For example, the use of poorly validated methods or statistics, wrong discovery strategy, confounding factors and many biases at various stages of the discovery process (patients’ recruitment, sample collection or analysis, etc.) [6–9].
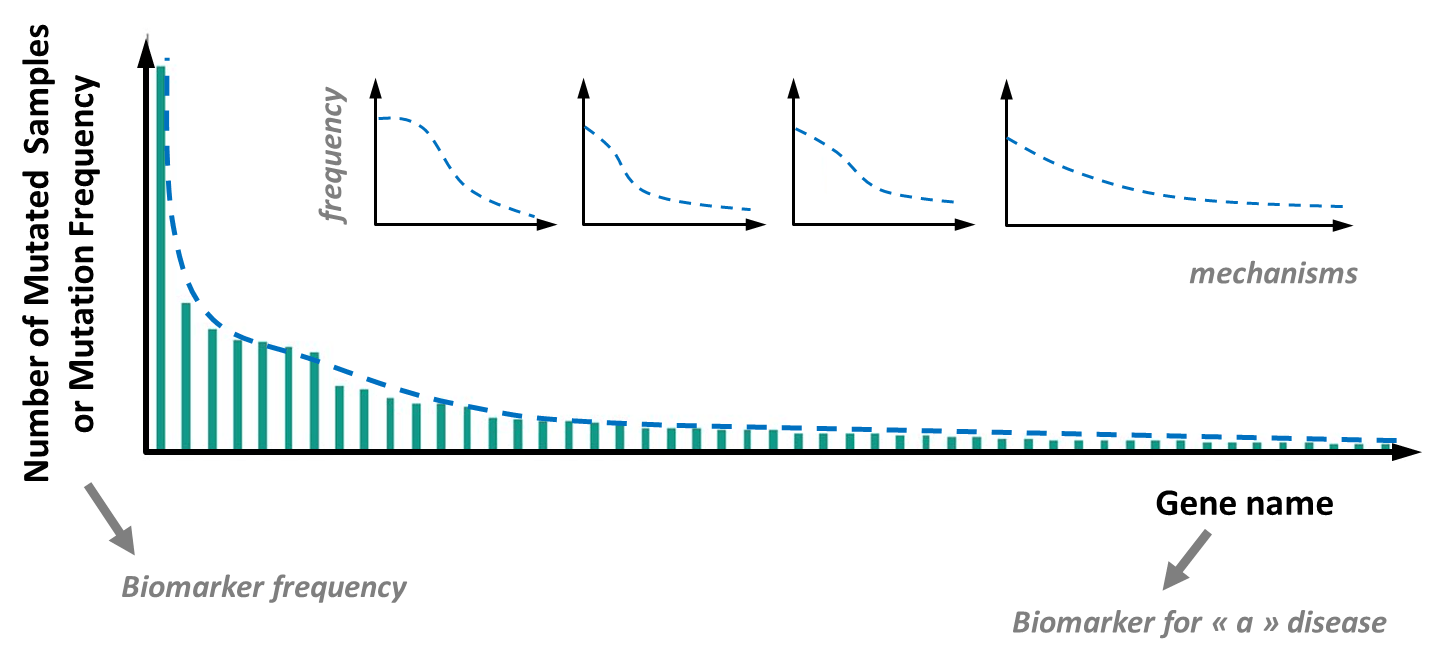
A new explanation of this ‘hangover’ takes its roots in a more complete integration of the living system complexity and the huge diversity of disease origins. This new comprehension of countless disease sub-categories and sub-mechanisms, can be more easily captured with recent data in cancer research, in cancer diagnosis and with the distribution of DNA mutation frequencies observed in the genome of tumor cells [10,11]. Even gene mutations in a particular gene are very diverse, with the possibility of missense or nonsense mutations, insertions or deletions (indels), multiplications, etc. This adds layers of complexity and underscores the need for global outlooks rather than restrictive targeted determinations [10,11].
This distribution of DNA mutation frequencies obeys a Power or Pareto’s law and is also called long tail distribution (see Figure 2). It reveals that even if a few DNA mutations are frequent in e.g. a tumor type, there are a countless number of mutations with intermediate, low or very low frequencies but with a potentially high degree of significance for the tumor behavior, diagnosis and anticancer treatment [10–12].
Step by step, cancer scientists have understood the huge genetic diversity of tumors to such an extent that, today, it is recommended to sequence the tumor genome (usually the coding part called the exome), or at least the hundreds of genes involved in cell growth, division, apoptosis, migration, aggressiveness etc., in order to deduce which treatment should be given to the patient [10,11]. Therefore, because it encompasses a large variety of possibilities, tumor genome sequencing is untargeted (even if only a small part of the genome is actually sequenced). Cancer diagnostics needs a global genetic determination of the tumor.
This comprehensive approach to tumor/cancer diagnosis should be more widely extrapolated to human diseases. Here also, the genome variability in the cells and tissues of a person has recently been reported to be much more important than previously thought [13]. The genome diversity between cells and tissues, is called mosaicism and is more the rule than the exception. This variability teaches us that there are many potential biomarkers with low or even very low frequencies (low sensitivity biomarkers) but with very high significance for diagnosis. Thus, it is recommended to record more global screening analyses than classical targeted determinations on a few biomarkers only.
The untargeted screening analysis is feasible routinely in a clinical lab [14] and will be needed to avoid misdiagnosis, a problem that is increasingly observed by health authorities [15,16]. Indeed, behind similar symptoms of a disease, lots of different mechanisms can be involved, suggesting that, like for tumors, there are many, many subtypes of the same disease and many low frequency biomarkers to search for (see Figure 2). Untargeted LC–HRMS approaches in clinical labs will become increasingly popular in cases where canonical approaches and targeted determinations fail to understand the mechanisms behind pain and symptoms.
Soon medical doctors will be aware of new LC–HRMS capabilities to get more global biomedical information obtained from untargeted diagnostic screening analysis [14]. Clinicians could ask for this untargeted outlook especially for non-responder patients. Indeed, there are many patients, who desperately look for a more accurate diagnosis and better treatment with greater levels of personalization.
In this context, bioanalysts working in clinical routine labs should look forward to performing such untargeted analysis and global determinations by high-resolution full scan or data-independent-analysis (e.g. MSSWATH). The path to look at disease diversity with the appropriate screening tools, such as LC–HRMS, is at its beginning. Medical doctors and bioanalysts should see the interest of untargeted diagnostic screening to provide more personalized diagnosis for better and cheaper healthcare.
- www.marketsandmarkets.com: Mass spectrometry market by application (pharmaceuticals, biotechnology, environmental testing), platform (single mass spectrometry (quadrupole, TOF & ion trap), hybrid mass spectrometry (triple quadrupole, QTOF & FTMS) global forecast to 2022. © 2017
- Rochat B. From targeted quantification to untargeted metabolomics: Why LC-high-resolution-MS will become a key instrument in clinical labs. Trends Analyt. Chem. 81, 151–164 (2016).
- Grund B, Marvin L, Rochat B. Quantitative performance of a quadrupole-orbitrap-MS in targeted LC–MS determinations of small molecules. Pharm. Biomed. Anal. 124, 48–56 (2016).
- Fedorova G, Randak T, Lindberg RH et al. Comparison of the quantitative performance of a Q-Exactive high-resolution mass spectrometer with that of a triple quadrupole tandem mass spectrometer for the analysis of illicit drugs in wastewater. Rapid Commun. Mass Spectrom. 27, 1751–1762 (2013).
- Rochat, E. Kottelat, J. McMullen. The future key role of LC-high-resolution-MS analyses in clinical laboratories: a focus on quantification. Bioanalysis, 4, 2939–2958, (2012).
- Ioannidis JPA, Bossuyt PMM. Waste, leaks, and failures in the biomarker pipeline. Chem. 63, 963–972, (2017).
- van Gool AJ, Bietrix F, Caldenhoven E et al. Bridging the translational innovation gap through good biomarker practice. Rev. Drug Discov.16, 587–588, (2017).
- Diamandis EP. The failure of protein cancer biomarkers to reach the clinic: why, and what can be done to address the problem? BMC Med. 10, 87 (2012).
- Drucker E, Krapfenbauer K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA J. 4,7 (2013).
- Hartmaier RJ, Albacker LA, Chmielecki J et al. High-throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res. 77, 2464–2475 (2017).
- Chang MT, Bhattarai TS, Schram AM et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 8, 174–183 (2018).
- Diamandis EP. A repository for ‘rare’ tumor markers? Chem. Lab. Med. 52, 795–797 (2014).
- Forsberg LA, Gisselsson D, Dumanski JP. Mosaicism in health and disease – clones picking up speed. Rev. Genet. 18, 128–142 (2017).
- Rochat B, Mohamed R, Sottas P.E. Untargeted diagnostic screening by LC–HRMS: a feasibility study for clinical laboratories. Metabolites 15, 8 (2018).
- www.nationalacademies.org/hmd/~/media/Files/Report%20Files/2015/Improving-Diagnosis/DiagnosticError_ReportBrief.pdf
- James JT. A new, evidence-based estimate of patient harms associated with hospital care. Patient Saf. 9, 122–128 (2013).





