Key challenges in sample preparation for peptide bioanalysis: a podcast with Mary Lame

https://soundcloud.com/bioanalysis-zone/key-challenges-in-sample-preparation-for-peptide-bioanalysis-a-podcast-with-mary-lame/s-fSyBK
In this interview, we talk to Mary Lame, Principal Applications Scientist who supports discovery bioanalysis for both large and small molecules, providing application development and support with Waters Corporation (Milford, MA, USA). Mary will discuss the key challenges scientists face developing robust peptide bioanalytical assays and introduces Waters approach for large molecule bioanalytical quantification method development including: pre-analytical sample preparation, as well as offering tips & tricks to help avoid the most common pitfalls working with peptides to help improve peptide assay performance and to streamline assay development. She also discusses how to enable highly sensitive and robust quantification of biomolecules from complex biomatrices.
1Could you brief introduce yourself?
Hi there and thank you for the opportunity to speak today. As stated, my name is Mary Lame, and I am a Principal Applications Scientist and Manager in the Scientific Operations group with Waters Corporation based in Milford, MA here in the United States. In this role, I assist in the research, development, and commercialization of Waters instrumentation and consumables, highlighting system solutions for the bioanalytical market. I began my bioanalytical career in 1997 with the global Pharmaceutical company, Pfizer (NC, USA), where I supported R&D efforts in the Drug Metabolism group, performing various in vivo and in vitro DMPK and biomarker studies and developing bioanalytical assays in support drug discovery across many disease areas, including: antibacterials, oncology, CNS and CVMED. I joined Waters in 2011 with the primary role to support discovery bioanalysis, developing methods for drug molecules, both large and small and providing customer and in-house training on these topics. I am responsible for end-to-end bioanalytical assay method development, which includes mass spectrometry, liquid chromatography, sample preparation and automated workflows.
2Could you provide an introduction into peptide bioanalysis – the what, why, how?
So let’s talk about peptide and proteins. Over the past decade, there has been a significant shift towards a greater percentage of biologics in pharmaceutical pipelines due to their improved efficacy and reduced side-effects. With this shift in large molecule therapeutics, the industry finds itself in the middle of patent expiry for many of the critical biologics. For example, the therapeutic peptide drugs Byetta, Forteo, and many insulin analogues like Lantus and Levimar. For protein-based drug therapeutics, we are talking about monoclonal antibody IgG based proteins, like Trastuzumab (Herceptin) and Infliximab (Remicade). This shift in large molecule biotherapies has resulted in an increased focus on their quantification in bioanalytical labs, innovator pharma, CROs, as well as biomarker research labs. With their increased size and structural complexity, compared to small molecule therapies, method development can be quite a daunting task for traditional highly skilled small molecule scientists, or even the novice.
To truly understand the therapeutic potential of peptides the right bioanalytical strategy and techniques must be employed and thus is the focus of today’s podcast. Over the next few minutes I will discuss and provide you with the basic tools/strategies and knowledge one needs for successful bioanalysis of peptides, with the ultimate goal of easing the transition from working on small molecules to large molecules. Today I want to discuss the challenges that face bioanalytical scientists when tasked with developing sensitive and robust LC–MS based methods for the analysis of large molecules, peptides and proteins from biological matrices, such as plasma, urine, CSF, etc.
3What are some of the biggest challenges in peptide sample preparation for LC–MS bioanalysis?
Great question, as we have seen the number of therapeutic biologic modalities increase in use over the past decade, the need for analysis of these biologics to support drug discovery and development, specifically when supporting ADME/DMPK studies has dramatically increased too. And this has brought new challenges to the typical small molecule bioanalytical laboratories. Specifically, new bioanalytical methods must be developed that are sensitive, selective and robust. These would ideally meet current regulatory criteria as well.
Successfully quantifying proteins and peptides with LC–MS is often more challenging than for small molecule therapeutics or modalities. Due to their large and complex structure, development of sensitive and robust peptide/protein analytical methods often requires more expertise and perhaps more method development due to the variety of challenges often encountered with these molecules. Because of their size, charge state distribution and complex MS/MS fragmentation of biomolecules, like peptides and proteins, sensitivity by mass spectrometry may be lower than typical small molecules, often necessitating greater LC and sample preparation method development. To gain better sensitivity and selectivity often requiring sample concentration and improved sample extraction techniques beyond basic protein precipitation – as a means of increasing overall method sensitivity and selectivity.
Specifically, in my experience developing end-to-end assays for these biomolecules, the challenges start from the pre-analytical handling and carry through to the sample extraction, chromatographic and MS method development. The typical challenges we often encounter are issues with maintaining solubility throughout sample preparation and analysis, effectively disrupting protein binding of the biomolecules from the biomatrices, non-specific binding (or adsorption) of these molecule to any surface it comes into contact with, which most almost always results in loss of analyte and higher variability of assay performance. Normally because these biomolecules, endogenous or therapeutic, are derived from 20 finite amino acids, we often encounter more matrix interferences, thus we need to achieve higher assay specificity. Lastly, low analyte recovery, which is the most common symptom of failure to successfully disrupt binding either non-specific or protein binding. When we wrap this all together failure to successfully deal with or overcome these challenges often leads to less than ideal analytical performance of the assays: lack of sensitivity, limited specificity, lack of robustness or higher variability (CVs or RSDs).
4Why should you be concerned about protein binding when trying to analyze peptides?
Well when thinking specifically about sample preparation, failure to effectively disrupt biomolecule from the biomatrices, prior to solid phase extraction (SPE), will cause the protein bound peptides to pass through the sorbent, unretained during initial loading of the sample, due to size exclusion mechanisms.
For these reasons, in our lab we often spend time assessing various effective pre-treatment strategies such as dilution with strong acid like 4% phosphoric acid, or 5% ammonium hydroxide solution as a starting point. Or even becoming more aggressive for the more challenging, larger and hydrophobic peptides. Here I am talking about organic pre-treatment with 1:1 methanol or acetonitrile, or denaturation with guanidine, urea or sodium dodecyl sulfate (SDS) as effective means of effectively disrupting protein binding. If we successfully do so we are inherently improving our recovery of our analyte through the extraction (driving sensitivity).
5What is non-specific binding and how does it impact quantitative performance?
When we talk about non-specific binding or NSB we are referring to the propensity of the peptide or protein to stick to or tend to bind or adsorb to any of the surfaces it comes in contact with, such as glass, collection vessels, pipette tips, LC fluidics, etc. The lower the concentration the more analyte losses that can occur. Additionally, the challenge here is that it is hard to predict NSB as it is influenced by the nature and size of the peptide, the organic composition of the diluent or solution the biomolecule is in, as well as concentration. So naturally, if the analyte is subject to NSB, we often find that our overall assays suffer. While we talk about the 5 key challenges, the symptoms we often see are poor sensitivity, higher LODs (due to the loss), chromatographic issues (poor peak shape, or none at all), non-linearity, particularly at the low end of a calibration curve and irreproducible chromatographic or quantitative performance.
6What strategies can you take to reduce non-specific binding?
Well first we want to consider solubility, ensuring that the biomolecule from the very start of initial solubilization from powder through to sample extraction and LC–MS analysis will minimize or lessen the severity of NSB. For this reason, we often recommend limiting organic concentrations of no more than 75%, as peptides will often precipitate in all organic, and use of organic modifiers like acid or base (anywhere from 1–10%) to promote and maintain solubility. This becomes especially important in SPE sample preparation.
Just as important as solubility so is the choice of collection vessels we use. Whether working with neat standard solution or low concentrations of extracted samples, the choice of collection vessel, which does not contribute to loss of the peptide via the NSB mechanism, is critical. Here we are talking about clean neat standards or processed samples that are highly purified and devoid of protein or other components that could protect the biomolecule from sticking to the container and being lost.
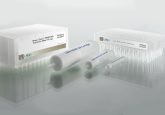
Part of our strategy here is to use low binding consumables. For us this problem of NSB is managed with the use of QuanRecovery vials and 96-well plates. These plates and vials are specifically designed for LC–MS analysis to reduce sample losses for biomolecules due to ionic interactions and non-specific binding with the sample container. They are provided in autosampler ready formats with low residual sample volumes for maximum sample utilization. Using low bind consumables ensures that we minimize loss from auto samplers, ensuring high recovery and repeatability of our assays.
7How do you isolate and clean-up concentrated peptides from blood/plasma?
While the most widely accepted bioanalytical extraction techniques are Protein Precipitation (PPT) with organic solvents and liquid–liquid extraction, these are often not sufficient for sample clean-up in terms of achieving recovery or reproducibility. And naturally once you start to gain more understanding of peptides and proteins, it becomes apparent this is often due to issues we have already discussed: poor recovery of PPT due to the peptide along with other proteins from the matrix being precipiated.
Thus, solid phase extraction using reversed phase and mixed mode then offers additional benefits. Allowing one to effectively and selectively purify the peptide from the matrices with high recovery (often greater than 75%) with high selectivity. Additionally, when one uses mixed mode extraction (use of cation or anion exchange) we can now add greater selectivity beyond RP SPE. And it adds orthogonality to the overall assay with the sample preparation performed in the mixed mode and the LC performed in the RP. Additionally, we can leverage the use of the micro elution format allows us to concentrate the sample directly eluting for extraction in as little as 25uL, without the need to evaporate and reconstitute, minimize losses of the analyte due to adsorption on evaporation or dry down of the sample or poor solubility when reconstitution.
8How can you prevent low recovery?
Well as we have gone over today, much of preventing loss or low recovery of peptides in a bioanalytical workflow starts with an appreciation for the key challenges: paying attention to the solubility of your peptides to avoid losses due to NSB. So you can use a mix or organic/aqueous solutions containing modifiers like formic acid or ammonium hydroxide to ensure proper ionization and maintain solubility. Also, ensuring proper pre-treatment, which effectively disrupts protein binding to fully recover your peptide from your biomatrices. Use of mixed mode solid phase extraction in the micro elution format can also be used to ensure high analyte recovery and sample concentration to improve selectivity and detection limits, and lastly using proper sample collection vessels like QuanRecovery vials and plates, which effectively and reliably reduce sample losses for biomolecules due to ionic interactions and non-specific binding with the sample container.
Our expert opinion collection provides you with in-depth articles written by authors from across the field of bioanalysis. Our expert opinions are perfect for those wanting a comprehensive, written review of a topic or looking for perspective pieces from our regular contributors.
See an article that catches your eye? Read any of our articles below for free.