Chapter 3: Proteomic approaches to biomarker discovery for prostate, bladder and kidney cancers
Doi: 10.4155/FSEB2013.13.32
Authors
Steven L. Wood*
Department of Oncology and Metabolism
The Medical School
University of Sheffield
Beech Hill Road, Sheffield
S10 2RX
Janet E. Brown
Academic Unit of Clinical Oncology
Broomcross Building
Weston Park Hospital, Sheffield
S10 2SJ
*Corresponding author
Tel: +114-2713167
Fax: +114-2713314
e-mail: [email protected]
About the Authors
Steven L. Wood
Steven Wood is a Senior Research Fellow within the bone-oncology and bone biomarkers group at the University of Sheffield. His research interests are focused on the proteomic analysis of bio-fluids for the identification of disease-associated biomarkers and the subsequent validation and clinical translation of this research.
Janet E. Brown
Janet Brown is Professor of Medical Oncology at the University of Sheffield, with specialist interests in uro-oncology and breast cancer, focusing on the effects of cancer on the skeleton. She is involved in a wide range of clinical trials in prostate, renal and breast cancer and in translational research for the development of biomarkers, leading to improved treatment strategies for these malignancies.
Proteomic approaches to biomarker discovery for prostate, bladder and kidney cancers
List of common abbreviations:
PCa = Prostate Cancer; BCa = Bladder Cancer; RCC = Renal Cell Carcinoma; MRM = Multiple Reaction Monitoring; MS = Mass-Spectrometry; 2D-DIGE = 2-Dimensional Fluorescence Difference Gel Electrophoresis; PTM = Post-Translational Modification; MALDI = Matrix-assisted laser desorption/ionisation; PSA = Prostate Specific Antigen; SWATH-MS = Signal Width Adjusted for All Theoretical Precursors-MS; MudPIT = Multi-Dimensional Protein Identification Technology; NMP22 = Nuclear Matrix Protein-22; iTRAQ = Isobaric Tagging for Relative and Absolute Quantification; SISCAPA = Stable-Isotope-Standards-And-Capture-by Anti-Peptide-Antibodies, NMIBC = Non-Muscle-Invasive-Bladder-Cancer; 2D-LC-MSMS = Two Dimensional Liquid Chromatography Tandem Mass Spectrometry; ELISA = Enzyme Linked ImmunoSorbent Assay; LC = liquid Chromatography; MSMS = Tandem Mass spectrometry; TOF = Time Of Flight; TOF-TOF = Tandem Time Of Flight; FT-ICR = Fourier Transform Ion-Cyclotron Resonance; HIF-1α = hypoxia-Inducible Factor-1α; EMT = Epithelial to Mesenchymal Transition; SDS-PAGE = Sodium Dodecyl Sulphate Poly-Acrylamide Gel Electrophoresis; gel-LC-MSMS = SDS-PAGE Gel – Liquid Chromatography – Tandem Mass Spectrometry analysis; EPS = Expressed Prostatic Secretion; SRM = Selected Reaction Monitoring; CE-MS = Capillary Electrophoresis Mass-Spectrometry; SELDI = Surface-enhanced laser desorption/ionisation; MALDI-TOF-MS = Matrix Assisted Laser Desorption and ionization Time Of Flight Mass Spectrometry, EGFR = Epidermal Growth Factor Receptor, EpCAM = Epithelial Cell Adhesion Molecule, MUC3 = intestinal mucin, PGA3 = Pepsinogen-3 Group 1, β2M = β-2 microglobulin.
Abstract
There is a pressing requirement for biomarkers for the major urological cancers, enabling earlier disease diagnosis, detection of malignant progression and the targeting of drug treatments. Proteomic analysis of urine as well as serum/plasma enables the identification of proteins which alter in response to cancer either in terms of expression level or post-translational modification. Improvements in protein fractionation and modern mass-spectrometric methods have enabled proteomics to deal with the challenges presented by these complex bio-fluids. This chapter details the challenges associated with proteomic analysis of bio-fluids, the recent progress in identifying potential biomarkers within the major urological cancers and outlines the steps necessary to translate these preliminary findings into eventual clinical application.
Introduction: Requirement for biomarkers within urological cancers
Cancers of the prostate, bladder and kidney are major causes of morbidity and mortality. These malignancies each present diverse clinical challenges, however a common requirement is for biomarkers enabling earlier diagnosis enabling more effective treatment before advanced tumour spread. There is also an additional requirement for biomarkers providing prognostic information aiding personalized medicine initiatives via detection of tumour recurrence and detection of drug -response profiles.
Prostate Cancer
Prostate cancer (PCa) is the second highest cause of cancer death in men and in 2008 there were 899,102 cases diagnosed worldwide [1]. Although the incidence of this cancer has increased, the mortality rate from PCa has recently decreased, largely due to improved diagnostic procedures. One of the most widely used screening procedures for PCa is serum prostate specific antigen (PSA) testing which measures the level of kallikrein-3, a glycoprotein enzyme produced by the prostate gland. Use of PSA testing has improved the early detection of PCa, however there are a number of limitations of this test. PSA levels can be proportional to the size of the prostate gland in some individuals and this varies between patients and serum PSA-levels are also known to increase with age [2]. As a consequence there is no accepted upper or lower limit cut-off for PSA. An age-specific cut-off of < 4ng/ml serum PSA has been accepted in most countries however patients with serum PSA-levels < 4ng/ml can still harbour PCa [3]. Attempts to improve the PSA test by measuring the ratio of free: serum-protein-bound-PSA, the rate (or velocity) of PSA-level increase or the measurement of specific PSA-isoforms have improved the diagnostic utility of the PSA assay, however increases in serum PSA can also arise due to benign conditions and therefore more sensitive and selective biomarker assays of PCa are still required. There is also a pressing need for biomarkers which can differentiate PCa from other non-malignant conditions of the prostate such as Benign Prostatic Hyperplasia (BPH) and High Grade Prostatic Intraepithelial Neoplasia (HGPIN).
Biomarkers are required for the detection of several transitions during PCA-development. In one developmental pathway PCa can progress from an early androgen-sensitive form to a castration-resistant form in cases in which anti-androgen therapy is utilised. An additional feature of PCa is its ability to metastasize to distant sites including bone and this is a principal cause of PCa-associated morbidity which can occur independently of the development of androgen-insensitivity. The ability to detect or predict these transitions at an early stage would enable a more timely consideration of appropriate therapeutic approaches.
Biomarkers of PCa are required to improve the sensitivity and specificity of current PSA-based testing as well as to provide prognostic information. The transition from an androgen-sensitive form of the disease to castration-resistance is a key step within PCa development and biomarkers enabling prediction/early diagnosis of this development will assist PCa-treatment.
Bladder Cancer
Bladder cancer (BCa) is the ninth most common solid tumour worldwide with 382,600 new diagnoses of BCa made worldwide in 2008 representing the fourth most common malignancy in men and the eighth most common in women [1]. Whilst early diagnosis biomarkers of BCa are required one of the most pressing clinical problems within BCa is the need to monitor patients for recurrence, which costs $3.4 billion annually within the USA. Several tests are currently used to detect BCa including BTA-STAT® (Polymedco, USA); BTA-TRAK® (Polymedco, USA); ImmunoCyt™ (Sanofi, Pasteur, Canada); Nuclear-Matrix Protein-22-NMP22®, BladderChek® and UroVysion® kits- all of which are FDA approved. Nevertheless they have a limited ability to detect NonMuscle-Invasive-Bladder-Cancer (NMIBC) and they are not applied in all countries. In view of these limitations of current markers there is a pressing need for further biomolecules to form the basis of future tests.
BCa is the ninth most common solid tumour worldwide. Due to the high incidence of tumour recurrence frequent patient testing by invasive cystoscopy is required. Non-invasive markers of tumour-recurrence and, in particular, non-muscle-invasive bladder cancer are therefore required.
Kidney Cancer
Renal Cell Carcinoma (RCC) is a malignancy arising from the proximal tubular epithelium of the kidney which is responsible for ~2% of all cancer deaths within western countries. RCC accounts for 2-3% of all malignancies diagnosed and, globally 273,518 new cases were reported in 2008 [1]. The most common form of RCC is clear cell carcinoma which is commonly diagnosed at a late stage for which the survival rate is lower (the 5-year survival rate for patients with metastatic RCC is 5% compared to 50-95% for patients with localized disease). RCC is relatively insensitive to radiotherapy and conventional chemotherapies and whilst targeted therapies (such as tyrosine-kinase inhibitors and mTOR inhibitors) are applied in patient management most patients eventually develop drug resistance. Biomarkers which can predict drug response or enable early diagnosis would therefore greatly assist the treatment of RCC.
RCC is frequently diagnosed at a late stage and is insensitive to currently applied therapies. Biomarkers are required which will enable the prediction of drug-response, facilitating personalized medicine initiatives.
Challenges in the proteomic analysis of serum/plasma and urine in the discovery of biomarkers for urological cancers
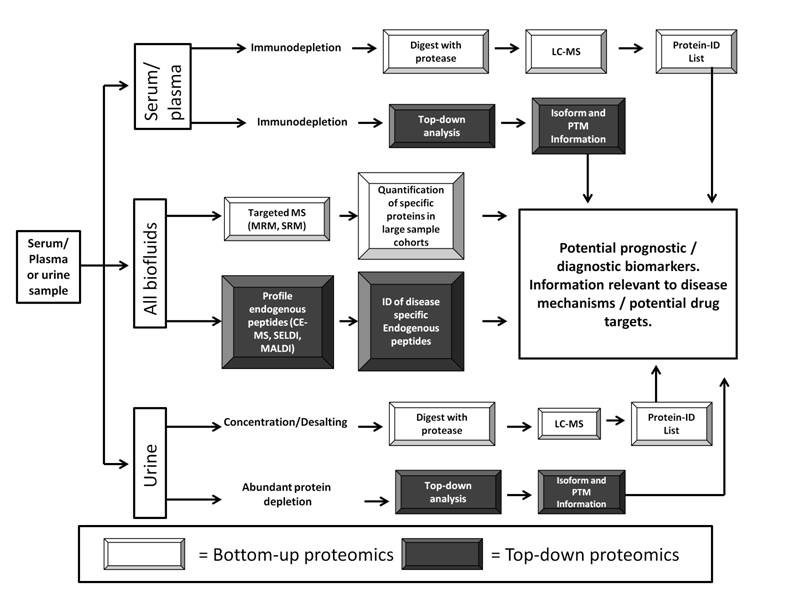
The biofluids with greatest potential for discovery of biomarkers within urological cancers are serum/plasma and urine. Both can be sampled non-invasively and contain proteins produced both by urological tumours as well as proteins altered in response to the presence of the tumour and are therefore potential biomarker repositories. Both of these biofluids can be analyzed by “top-down” and “bottom-up” proteomic analysis. In terms of proteomic analyses urine and plasma/serum present diverse challenges in terms of biomarker discovery. “Top-down Proteomics” refers to the isolation and quantitative analysis of intact proteins and peptides from body fluids providing information relevant to disease-specific post-translational modifications, proteolysis and alternative splicing of genes. Techniques include: 2D-DIGE [4] and peptide profiling via MALDI-TOF-MS or SELDI-TOF-MS [5]. Top-down techniques such as SELDI-TOF-MS suffer from several technical limitations including poor technical reproducibility and difficulty in terms of identifying the proteins responsible for the biomarker peaks observed. “Bottom-up proteomics” refers to the identification and quantification of proteins following their chemical or enzymatic digestion followed by sequencing or mass determination of the resulting peptides. Examples of bottom-up techniques include: iTRAQ [6] and TMT [7] based quantitative proteomic analysis. Identification of differential expression of protein isoforms by bottom-up methods is less direct than by top-down approaches but it can provide high sensitivity and high sample throughput enabling wide proteomic coverage.
Analysis of the urinary proteome
Urine contains proteins either secreted or directly shed from the bladder, prostate and kidney, with healthy adults secreting 1-4 g of peptide and 0.15g of protein per day within urine. The main challenges within proteomic analysis of urine include high inter-individual and intra-individual variability, relatively high sample salt concentration, a preponderance of a small number of highly abundant proteins (eg. uromodulin) and the difficulty of data normalization to produce robust comparative data. Several approaches have been utilized to overcome these challenges (see Figure 1). In order to minimise technical variability within urine samples, different protocols recommend standardized collection procedures (eg. first void urine for PCa studies where prostatic secreted proteins and peptides are detected) and data normalization to “housekeeping” peptides with constant expression levels (i.e. nine highly abundant peptides derived from collagen). Sample preparation for most proteomic projects involves urine concentration and desalting, although careful assessment of the effects of these steps upon sample variability is required. Sample pre-fractionation is a key step within the analysis of urine and may involve the selective enrichment for sub-cellular components such as exosomes (see section 5.0 below), or protein fractionation (e.g. glycoprotein enrichment).
Urine is a source of secreted and directly shed proteins arising from the endothelial cells lining the urogenital tract. As such it is a potential source of non-invasive biomarkers. The main challenges of urinary-proteomics are high sample variability, low protein concentration and high salt concentration.
Analysis of the serum/plasma proteome
Serum/plasma presents different challenges within biomarker discovery. The large dynamic range of protein expression within serum/plasma means that the top 22 most abundant proteins account for 99% of the total protein mass. These high abundance proteins can mask lower abundance biomarker proteins requiring either immunodepletion and/or extensive fractionation to maximise the number of proteins and overall sensitivity of the method employed (see Figure 1). Multi-dimensional LC-separation of serum using electrostatic-repulsion hydrophilic-interaction liquid chromatography (ERLIC), high-PH reverse-phase-LC and low-pH nano-LC-MSMS recently enabled the identification of 1088 peptides within serum [8]. The Human Proteome Organization (HUPO) consortium database currently lists 1929 non-redundant protein sequences, a combined list from the studies of ~30 different laboratories [9]. Despite these advances however proteomic analysis of serum/plasma samples is still challenging. A summary of the major methods within proteomic profiling of biofluids within urological cancers is provided by Figure 1.
Serum / plasma is a potential non-invasively sampled source of biomarkers containing proteins representing the physiological state of all major tissues. Proteomic analysis of serum / plasma is challenging owing to the wide-dynamic range of protein expression and high sample complexity. Despite these challenges progress in the field of biomarker discovery for urological cancers is being made.
Urinary biomarkers
Prostate Cancer
In order to increase the sensitivity of urine-based proteomic workflows to detect prostate derived proteins, studies have utilised urine samples collected post digital-rectal examination (DRE)-a process which increases the concentration of prostate-released secretions into urine. A recent MudPIT analysis of post-DRE urine from biopsy-negative non-cancer patients recently identified 1022 unique urinary proteins [10] and further analysis of pre- and post-DRE urine samples from PCa and non-PCa sources identified a 49 protein panel. Seven of these proteins (transglutaminase – 4-TGM4, lactotransferrin – LTF, alanyl-membrane aminopeptidase – ANPEP, membrane metalloendopeptidase – MME, tissue inhibitor of metalloproteinase – 1-TIMP1, Parkinson protein – 7-PARK-7 and 14-3-3σ) being validated by western blotting within an independent test set of DRE-urine samples from patients with and without biochemical evidence of recurrence, as well as by targeted SRM-MS within post-DRE-urine samples from men with extra-capsular and organ-confined PCa tumours [10]. In an independent study a protein panel was also discovered by MudPIT analysis of post-DRE urine samples from patients with organ-confined and extra-capsular PCa [11]. TGM4, PARK7 and MME were proteins discovered as potential biomarkers within both these studies. PARK7 has been identified as a mediator of both innate immunity response via the Toll-like Receptor as well as playing a role within the regulation of HIF-1α-expression. TGM4 has been demonstrated to increase the cell-adhesion, invasiveness and EMT-properties of PCa cells in culture. Cancer cell invasion is known to involve differential protease activation for which MME could be one key mediator. Increased MME expression has been observed within PCa-tumour adjacent epithelium samples compared to histologically benign epithelium samples. The discovery of the altered levels of PARK7, TGM4 and MME proteins thus aids our understanding of PCa development as well as providing a potential diagnostic biomarker panel. A recently reported study using iTRAQ-based quantification of urine samples from patients with BPH and prostate cancer identified β-2-microglobulin (β2M), pepsinogen-3 group-1 (PGA3) and intestinal mucin (MUC3) as a potential diagnostic panel for prostate cancer with an area under curve (AUC) for the combined panel of 0.710, a diagnostic accuracy further improved to 0.812 by combination with PSA status [12].
Bladder Cancer
Abundant protein depletion within bladder cancer and control (hernia) urine samples followed by iTRAQ-based quantitative proteomics recently identified a panel of seven proteins diagnostic of BCa, with a combination of SAA4 and pro-EGF distinguishing BCa and hernia cases with AUC 0.80, p < 0.001 [13]. A combined transcriptomic / proteomic study utilised shotgun proteomics to discover the proteins present within the secreted fraction (“secretome”) of BCa cells in culture [14] with subsequent downstream analysis to identify which of these proteins were encoded be genes upregulated in BCa tumours vs. normal urothelium [14]. This combined analysis identified a series of biomarker candidates subsequently validated by ELISA within urine from BCa patients with varying stages of disease [14]. This study identified the inhibitor of the protease responsible for hepatocyte-growth factor activation (HAI-1) and the pro-angiogenic growth factor Midkine as potential BCa biomarkers, with a particular utility for muscle-invasive disease [14]. A recent shotgun proteomic analysis of the proteins released by a panel of bladder cancer cell-lines identified the soluble ectodomain of epidermal growth factor (EGFR) and epithelial cell adhesion molecule (EpCAM) as markers of cancer survival, and measurement of these two molecules within urine was found to predict bladder cancer specific survival with a prognostic value above that of standard clinical observations [15].
Kidney Cancer
To date no diagnostic biomarkers have been identified for kidney cancer within urine (nor indeed within plasma, tissue or cells). One approach used to facilitate targeted analysis of proteins within urine is the analysis of tumour-proximal fluids. A proximal fluid relevant to RCC is tumour-cyst fluid and reverse-phase-liquid chromatography-based separation of RCC cyst-fluid and MSMS analysis identified 14-3-3β/α as a potential RCC-biomarker [16]. Urinary 14-3-3β/α-levels were predictive of RCC with a ROC AUC of 0.8813 [16]. In another study which aimed to use proximal fluids to direct RCC-urine analysis, 2D-DIGE-based analysis of conditioned media from RCC and control immortalised cell-lines identified cathepsin-D as a potential prognostic/diagnostic biomarker for RCC [17]. The study identifying cathepsin-D as a potential diagnostic/prognostic biomarker was initially performed by comparison of conditioned media from 4 RCC and 4 normal renal cultures and this observation was validated by ELISA within 239 patient derived urine samples (139 patients with RCC, 30 with benign kidney conditions and 60 healthy patient controls) [17]. The authors of this study recommend validation within a larger patient cohort however such a validation has not been reported to date.
Serum/Plasma Biomarkers
Although proteomic studies using urine as a potential biomarker source predominate due to the fact that this biofluid more directly samples shed/secreted proteins from the urogenital tract, serum/plasma has potential to provide a non-invasively sampled source of proteomic biomarkers for urological cancers. Sub-fractionation of serum/plasma to aid proteomic detection of biomarker proteins is a common feature of these studies.
Prostate Cancer
Top-down proteomic studies have been employed in the discovery of serum/plasma-based PCa-biomarkers. In one study using lectin-based glycoprotein-enrichment and 2D-DIGE analysis a biomarker panel of 6-different proteins were identified which could distinguish PCa from BPH (including complement C3-β-chain fragment, inter-alpha-trypsin inhibitor heavy-chain 4-fragment, transthyretin, alpha-1-antitrypsin, and high-MW kininogen [18]).
The validation and clinical application of proteomic biomarkers is discussed in section 6.0 however mass-spectrometry also has a role in this important process. Quantification of the absolute levels of proteotypic peptides can provide an assay used to assess biomarker utility in large patient cohorts. Proteomic comparison of serum samples from PCa patients identified two potential marker peptides which could predict disease recurrence [19] including a truncated form of complement C4a and the N-terminal fragment of protein C-inhibitor (PCI). Use of a heavy isotopically labelled form of this proteotypic PCI-peptide within a SISCAPA-MALDI study confirmed the utility of this PCI-derived peptide for monitoring disease recurrence within a large cohort of patients receiving radiation and hormone-therapy.
Bottom-up analysis of PCa-serum samples using iTRAQ within patients with benign-prostatic hyperplasia, localized cancer with no signs of progression, PCa samples with evidence of progression and patients with bone-metastases enabled the quantification of 122 proteins with a 23 protein subgroup differing between metastatic and progressing but non-metastatic patients [6]. One of these proteins eukaryotic elongation factor-1-alpha (eEF1A1) was identified as being closely associated with PCa metastasis and immunostaining for eEF1A1 was observed within tumour-proximal osteoblasts within tissue sections [6].
Bladder Cancer
Serum-based profiling studies for the identification of BCa biomarkers have focussed upon the low-MW peptidome using techniques such as MALDI-TOF-MS. In a representative study a role for exoprotease-generated peptides derived from the ex-vivo coagulation and complement degradation pathways was identified in a MALDI-TOF-MS based study comparing patients with 3 different types of solid tumours and healthy controls [20]. These tight peptide clusters corresponded to 61 different peptide peaks [20]. These studies reveal the potential of the low-MW serum peptidome to provide a source of diagnostic markers for BCa.
Discovery of serum-based bladder cancer biomarkers has involved the use of both antibody and protein arrays which enable quantification of specific proteins using either immobilized capture antibodies (for antigen quantification) or immobilised proteins (for quantification of circulating antibodies). In a small patient cohort protein array-based study using 12 BCa patient serum samples and 10 control serum samples 171 proteins were observed to be differentially expressed within BCa, including the proteins dynamin and clusterin [21]. Validation of these proteins by immunohistochemistry using 289 patient-derived TMAs revealed that dynamin and clusterin may be useful for tumour staging and the diagnosis of muscle-invasive disease [21]. In a separate study use of antibody-arrays within serum-profiling identified a protein-profile which could distinguish BCa from control patients with 96.5% specificity and 89.2% sensitivity [20]. Within this study some of the top-ranked altered proteins associated with BCa (i.e. c-Met) were statistically significantly associated with pathological stage, tumour grade and survival [20].
Kidney Cancer
A wide-range of diverse proteomic techniques have been employed in the identification of serum/plasma-based biomarkers for RCC. Within a comparative study of serum-derived peptides from RCC and control samples using reverse-phase-magnetic bead enrichment a panel of 200 peptides derived from 32 -different proteins were identified discriminating RCC from control samples [22]. This MALDI-TOF-based study was further extended by MALDI-TOF-TOF and MALDI-FT-ICR based sequencing, identifying human serum-deprivation response-protein-3, zyxin , serglycin and thymosin-like-3 protein as potential serum-based biomarker candidates for RCC. Application of a multiplexed immunoassay-based screen to plasma samples from a panel of healthy patients and those with benign tumours in comparison with kidney cancer patients in a training set identified a panel of three proteins nicotinamide-N-methyltransferase (NNMT), nonmetastatic cells 1 protein (NM23A) and L-plastin which could diagnose the presence of kidney cancer with 90% specificity and 95.7% sensitivity within a subsequent validation test cohort [23].
Exosomes as a potential biomarker-enriched sample source:
Biomarker identification projects within urological cancers have begun to focus upon the exosomal fraction of urine and plasma/serum. Exosomes are small (30-100 nm) secreted micro-vesicles which play an important role in cell-cell contact and the ability of cancer cells to reshape their environments. Exosomes are present within major biofluids (including serum/plasma and urine) and the study of urinary exosomal proteins offers the advantage of reduction in sample complexity combined with a focus on a specific and highly-relevant protein subclass – see Figure 1.
A comparative gel-LC-MSMS study of urinary exosomes isolated by differential centrifugation from RCC (n = 29) patients and healthy sex matched (n = 23) controls identified a panel of 10 proteins (including aquaporin-1, dickkopf-related protein-4 and carbonic anhydrase-IX) as differentially expressed within RCC [24]. The discovery of these differentially expressed exosomal proteins may assist in the development of a diagnostic biomarker test for RCC.
In a recent quantitative-proteomic study comparing urinary exosomes derived from BCa and hernia patients using dimethyl-labelling and sample multiplexing 29 proteins were quantified as differentially expressed [25]. Extension of this study by ELISA identified tumour-associated calcium-signal transducer-2 as a potential exosomal biomarker for BCa [25].
In some cases exosomes have been implicated in processes such as angiogenesis, tumour-invasion and tumour progression suggesting that differential protein profiling may identify potential drug targets. RCC-stem cell released exosomal vesicles have also been demonstrated to stimulate angiogenesis and the development of a pre-metastatic niche within the lung and thus exosomal profiling my eventually identify anti-metastatic drug targets. Proteomic studies of urinary exosomes from PCa, BCa and RCC have identified a series of potential biomarker proteins for these major urological cancers (see Table 1).
Table 1: Proteomic studies of exosomes within major urological cancers:
| Source Material | Method | Proteomic data-set | Reference |
| Prostate Cancer (PCa) | |||
| DRE-Urinary exosomes. | Shotgun proteomic – control and PCa pools, | ~900 proteins discovered. | [26] |
| PCa, PBH and healthy control serum. | Centrifugal enrichment- immunoassay analysis. | Elevated survivin levels within PCa patients compared to BPH and healthy controls. | [27] |
| PCa-derived vertebral metastases. | Tissue homogenization and centrifugation. | Twenty five proteins identified all potential angiogenic mediators. | [28] |
| Bladder Cancer (BCa) | |||
| BCa cell lines and BCa patient (urine). | Centrifugal isolation. | 353 proteins identified, 18 proteins validated. | [29] |
| BCa patients and control hernia patients. | Differential centrifugation and dimethyl-labelling. | 107 differentially expressed proteins identified as potential biomarkers between BCa and hernia – 2 taken forward for ELISA-based validation. | [25] |
| Kidney Cancer (RCC) | |||
| RCC patients and healthy controls. | Centrifugal isolation and gel-LC-MSMS. | 261 proteins identified (control patient urine), 186 proteins identified (RCC-patient urine). Specificity confirmed by comparison to BCa-derived urine. 10 differentially expressed proteins validated. | [24] |
Exosomes are 30-100nm secreted microvesicles which contain proteins (and also regulatory micro-RNAs) representing the physiological state of the host cell. As such exosomes are a potential biomarker repository. In the case of both serum/plasma and urine based studies exosomal enrichment reduces the problems arising from high-abundance proteins within proteomic analysis of these bio-fluids.
Conclusions: Advancing proteomic biomarkers towards clinical application
Whilst clinical proteomics offers considerable promise to improve early diagnosis and patient care, within urological cancers to date the transition of proteomic-biomarkers into clinical tools has been slow. The Ova1® (Vermillion, USA) kit is the only example of a biomarker discovered by proteomics which is currently used in the clinic.
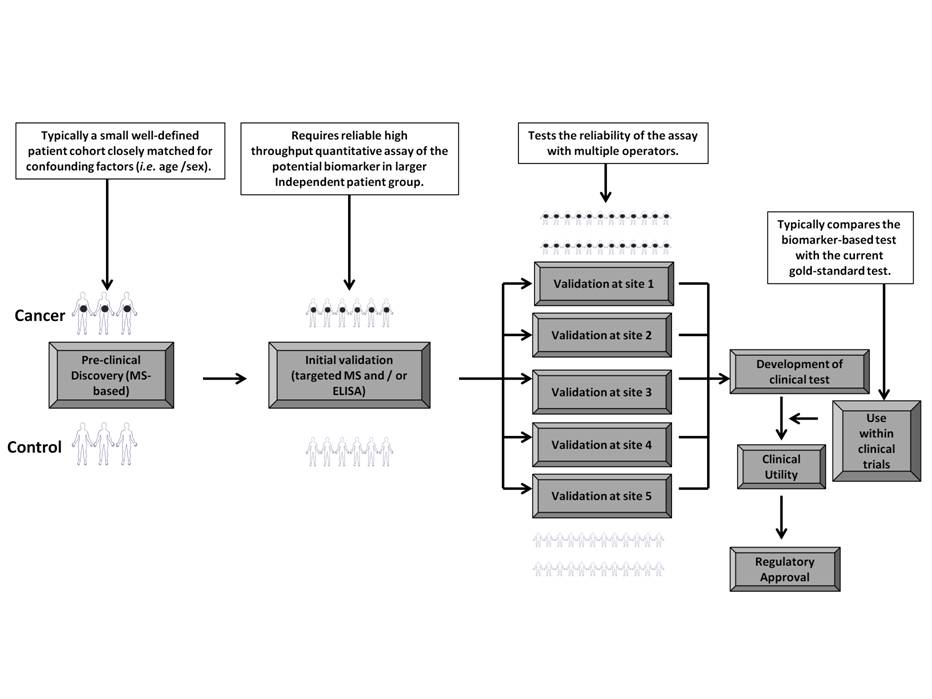
Several challenges have prevented the efficient translation of lab-based discoveries into the clinic. The standard approach for development of biomarker candidates into clinical tools involves the application of the originally discovered biomarker test within larger blinded test sets of patients controlled as closely as possible for confounding factors such as age and sex (see Figure 2). Many original biomarker reports are not confirmed upon repeat testing and this may reflect insufficient sample size within the original discovery experiments or biases introduced during patient selection. It is also essential to standardize sample collection and processing steps as proteins may not have sufficient stability within urine, thus being lost when time to sample processing is extended.
Efficient biomarker validation requires the development of suitably high-throughput quantitative assays for the protein in question [30]. Increasingly mass-spectrometric methods are being employed for high throughput targeted protein quantification. Recent techniques such as SWATH-MS and SISCAPA-assisted Multiple Reaction Monitoring (MRM) have begun to impact upon high throughput protein quantification within plasma/serum as well as within urine (MRM-based quantification of 63 different proteins within urine being recently reported [31]). These techniques are cost-efficient, have a low limit of detection, can be multiplexed to assay multiple proteins simultaneously and have high accuracy [32]. In a recent study targeted MS approaches were applied to 1000 different cancer-associated proteins within plasma and urine enabling detection of 182 proteins within plasma and 408 proteins within urine and the quantification of 34 differentially expressed proteins across a panel of 83 plasma samples [33]. There is hope that targeted MS approaches may bridge the gap between rate of discovery of potential biomarkers and their rate of subsequent validation.
When proteomics-based biomarkers progress through validation in an independent patient cohort, then subsequent multi-institutional validation can form the basis of clinically applicable biomarker tests. Owing to the often heterogeneous nature of urogenital cancers in terms of driver mutations, and the high intra- and inter-individual variability within urine samples there is a growing realisation that multi-protein marker panels may give better sensitivity/specificity for the clinical diagnosis of cancer, as well as providing potential prognostic information aiding patient treatment decisions. The ultimate clinical application of biomarker-based tests is usually an immunoassay kit which, in the case of the Ova1® FDA-approved test kit, is a slide-based immunoassay. The final developed assay-kit then requires regulatory approval following use in clinical trials proving the test’s robust applicability for informing treatment decisions within the cancer in question.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. No writing assistance was utilized in the production of this manuscript.
Executive summary:
- There is a pressing requirement for early diagnosis, prognostic and treatment-monitoring biomarkers for the major urological cancers (PCa, BCa and RCC),
- Urine and serum / plasma are potential sources of non-invasively sampled biomarkers for urological cancers. Urine in particular directly samples the major tissues of the urogenital tract,
- Proteomic analysis of each of these bio-fluids faces unique challenges, however developments in sample fractionation and MS have enabled identification of putative biomarkers for the major urological cancers,
- Exosomes are a potential source of biomarker proteins as they can be enriched relatively easily and they contain proteins representative of the physiological state of the tissues of origin. Exosome-derived biomarkers have begun to be identified for the major urological cancers,
- The translation of proteomic biomarkers into clinical application is a process involving demonstration of biomarker applicability within larger patient cohorts as well as the development of reliable biomarker assays based upon the proteins discovered by proteomic workflows.
Key Terms:
Urological cancers: Cancers that affect the tract responsible for the production of urine, namely prostate bladder and kidney cancer.
Mass-spectrometry: A term encompassing a wide range of analytical techniques and hardware platforms that all have the common feature of producing spectra of the masses of atoms/molecules within a sample.
Biomarkers: A biomarker is a measured characteristic which may be used as an indicator of some biological state or condition. In the context of proteomics a biomarker is a protein for which the change in expression or state correlates with presence of disease, the risk of developing the disease, the progression of a disease, or with the susceptibility of the disease to a given treatment
Exosomes: Exosomes are 30-100nm vesicles released by cells and present within body fluids and cell-culture media. They contain cell-derived proteins and nucleic acids and may be a potential source of disease-specific biomarkers.
Biofluids: Are body fluids. Within proteomics and urological cancers the predominant biofluids are plasma/serum and urine.
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer. Journal international du cancer, 127(12), 2893-2917 (2010).
- Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nature reviews. Cancer, 8(4), 268-278 (2008).
- Krumholtz JS, Carvalhal GF, Ramos CG et al. Prostate-specific antigen cutoff of 2.6 ng/mL for prostate cancer screening is associated with favorable pathologic tumor features. Urology, 60(3), 469-473; discussion 473-464 (2002).
- McNamara LE, Dalby MJ, Riehle MO, Burchmore R. Fluorescence two-dimensional difference gel electrophoresis for biomaterial applications. Journal of the Royal Society, Interface / the Royal Society, 7 Suppl 1, S107-118 (2010).
- Liu C. The application of SELDI-TOF-MS in clinical diagnosis of cancers. Journal of biomedicine & biotechnology, 2011, 245821 (2011).
- Rehman I, Evans CA, Glen A et al. iTRAQ identification of candidate serum biomarkers associated with metastatic progression of human prostate cancer. PloS one, 7(2), e30885 (2012).
- Raso C, Cosentino C, Gaspari M et al. Characterization of Breast Cancer Interstitial Fluids by TmT Labeling, LTQ-Orbitrap Velos Mass Spectrometry, and Pathway Analysis. J Proteome Res, (2012).
- Boichenko AP, Govorukhina N, van der Zee AG, Bischoff R. Multidimensional separation of tryptic peptides from human serum proteins using reversed-phase, strong cation exchange, weak anion exchange and fused-core fluorinated stationary phases. J Sep Sci, (2013).
- Farrah T, Deutsch EW, Omenn GS et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics, 10(9), M110 006353 (2011).
- Principe S, Kim Y, Fontana S et al. Identification of prostate-enriched proteins by in-depth proteomic analyses of expressed prostatic secretions in urine. J Proteome Res, 11(4), 2386-2396 (2012).
- Kim Y, Ignatchenko V, Yao CQ et al. Identification of differentially expressed proteins in direct expressed prostatic secretions of men with organ-confined versus extracapsular prostate cancer. Mol Cell Proteomics, 11(12), 1870-1884 (2012).
- Jedinak A, Curatolo A, Zurakowski D et al. Novel non-invasive biomarkers that distinguish between benign prostate hyperplasia and prostate cancer. BMC cancer, 15, 259 (2015).
- Chen CL, Lin TS, Tsai CH et al. Identification of potential bladder cancer markers in urine by abundant-protein depletion coupled with quantitative proteomics. J Proteomics, 85, 28-43 (2013).
- Shimwell NJ, Bryan RT, Wei W et al. Combined proteome and transcriptome analyses for the discovery of urinary biomarkers for urothelial carcinoma. Br J Cancer, 108(9), 1854-1861 (2013).
- Bryan RT, Regan HL, Pirrie SJ et al. Protein shedding in urothelial bladder cancer: prognostic implications of soluble urinary EGFR and EpCAM. British journal of cancer, 112(6), 1052-1058 (2015).
- Minamida S, Iwamura M, Kodera Y et al. 14-3-3 protein beta/alpha as a urinary biomarker for renal cell carcinoma: proteomic analysis of cyst fluid. Anal Bioanal Chem, 401(1), 245-252 (2011).
- Vasudev NS, Sim S, Cairns DA et al. Pre-operative urinary cathepsin D is associated with survival in patients with renal cell carcinoma. Br J Cancer, 101(7), 1175-1182 (2009).
- Jayapalan JJ, Ng KL, Razack AH, Hashim OH. Identification of potential complementary serum biomarkers to differentiate prostate cancer from benign prostatic hyperplasia using gel- and lectin-based proteomics analyses. Electrophoresis, 33(12), 1855-1862 (2012).
- Rosenzweig CN, Zhang Z, Sun X et al. Predicting prostate cancer biochemical recurrence using a panel of serum proteomic biomarkers. J Urol, 181(3), 1407-1414 (2009).
- Villanueva J, Shaffer DR, Philip J et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest, 116(1), 271-284 (2006).
- Orenes-Pinero E, Barderas R, Rico D et al. Serum and tissue profiling in bladder cancer combining protein and tissue arrays. J Proteome Res, 9(1), 164-173 (2010).
- Gianazza E, Chinello C, Mainini V et al. Alterations of the serum peptidome in renal cell carcinoma discriminating benign and malignant kidney tumors. J Proteomics, 76 Spec No., 125-140 (2012).
- Su Kim D, Choi YD, Moon M et al. Composite three-marker assay for early detection of kidney cancer. Cancer Epidemiol Biomarkers Prev, 22(3), 390-398 (2013).
- Raimondo F, Morosi L, Corbetta S et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst, 9(6), 1220-1233 (2013).
- Chen CL, Lai YF, Tang P et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J Proteome Res, 11(12), 5611-5629 (2012).
- Principe S, Jones EE, Kim Y et al. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics, 13, 1667-1671 (2013).
- Khan S, Jutzy JM, Valenzuela MM et al. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One, 7(10), e46737 (2012).
- Ronquist KG, Ronquist G, Larsson A, Carlsson L. Proteomic analysis of prostate cancer metastasis-derived prostasomes. Anticancer Res, 30(2), 285-290 (2010).
- Welton JL, Khanna S, Giles PJ et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics, 9(6), 1324-1338 (2010).
- Whiteaker JR, Lin C, Kennedy J et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol, 29(7), 625-634 (2011).
- Chen YT, Chen HW, Domanski D et al. Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. J Proteomics, 75(12), 3529-3545 (2012).
- Percy AJ, Chambers AG, Yang J, Hardie DB, Borchers CH. Advances in multiplexed MRM-based protein biomarker quantitation toward clinical utility. Biochim Biophys Acta, (2013).
- Huttenhain R, Soste M, Selevsek N et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med, 4(142), 142ra194 (2012).
Figure 1: Proteomic analysis workflow for discovery of bio-fluid-based biomarkers within urological cancers.
The two most commonly utilised body fluids within biomarker discovery within urological cancers are urine and plasma/serum. Both provide unique opportunities and challenges.
Urine is a potential source of non-invasively sampled biomarkers for urological cancers. Commonly applied pre-fractionation steps include depletion of abundant proteins (in particular uromodulin but also human serum albumin), combined with sample desalting/concentration, steps which are particularly useful for urine due to the high salt concentration of this body fluid. Further protein fractionation can improve the number of proteins identified within urine samples.
The proteomic analysis of serum / plasma samples derived from urological cancer patients can provide considerable information regarding altered proteins released into the circulation during disease-progression. Due to the preponderance of high-abundance proteins within serum / plasma immunodepletion is a common first step before either bottom-up analysis or the identification of altered protein isoforms or PTMs within top-down protein analysis.
Several techniques can be employed with both urine and serum/plasma (“All Biofluids” workflow) including targeted MS which have increasingly been used for biomarker development / validation and more recently biomarker discovery. The insights provided by plasma / serum proteomics can supplement the discoveries made by urine-based proteomics to provide a thorough description of the alterations in protein expression occurring within urological cancer. The profiling of endogenous peptides within bio-fluids also has considerable potential to detect cancer-related biomarkers.
Figure 2: Translation of MS-based proteomic markers into clinically-applicable biomarkers: A proposed pipeline. LC-MS-based proteomics has to date produced a considerable number of potential biomarkers for urological cancers, however their translation into clinical applicability requires a number of challenging steps. The sensitivity and specificity of the proposed biomarker (or increasingly the biomarker panel) has to be established within larger patient cohorts and biomarker validation also requires the demonstration of applicability of the biomarker within multiple assessment sites. This validation pipeline requires the development of robust, high-throughput, quantitative assays (typically ELISA but increasingly targeted-MS approaches are being used). Final application of the biomarker within patient treatment requires the development of clinical tests which can be used by non-lab based clinicians and their use within clinical trials (and comparison with current gold-standard tests) eventually forms part of the process leading to their regulatory approval.